reference to original doc: https://arxiv.org/pdf/2310.06552 Grok chat I had handy: https://grok.com/share/c2hhcmQtMg%3D%3D_5e2db47f-1b2c-44eb-92d4-7362a361e23f
tldr:
Assigning ICD codes is time consuming and requires a specialist, automating would save lots of money and error. LLMs already know lots about disease classification, using two prompt frameworks the paper uses the ICD tree structure to get better scores than state-of-the-art on several classification scores. Tested with Codi-Esp dataset, GPT 3.5 + 4, and LLama 2. Is useful to integrate into an existing healthcare system.
Meat and Bones 🍖
This paper is a great look at some applications of LLMs in positive tasks! We’re looking at a multi labelling classification problem for ICD (International Classification of Disease) codes, which are numerical codes that correspond to a given disease when presented with clinical notes. The interesting quality that this paper attempts to leverage is the hierarchical structure of these codes, using “off the shelf” LLMs to one-shot the automation.
Mentioned earlier that ICD codes are hierarchical, they essentially follow a tree like structure to become more specific as we step down layers:
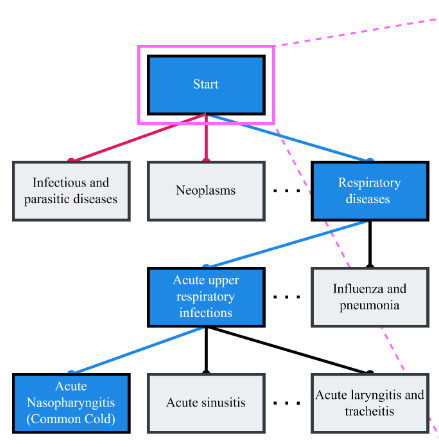
The sections of the tree are split like so:
- Chapters: Top-level categories based on broad disease groups or body systems (e.g., Chapter 10: Diseases of the Respiratory System).
- Blocks: Subdivisions within chapters, covering specific ranges of conditions (e.g., J00-J06: Acute Upper Respiratory Infections).
- Categories: More specific groups within blocks (e.g., J00: Acute Nasopharyngitis).
- Subcategories: Detailed codes within categories, often specifying variations or complications (e.g., J00.0: Acute Nasopharyngitis, Common Cold).
- Extensions: Additional digits for further specificity, such as laterality or severity (e.g., C63.2: Malignant Neoplasm of Scrotum).
The aim is to get to an extension with every assignment, but quite often this will not be the case owing to the nature of diagnosis and the multi variance that drives assignment. The strategy for this paper involved two prompt frameworks, task/profession framing and a guided tree search. What I found quite interesting was that the model temperature was set to 0, which I learnt was just a hyperparam surrounding the confidence models had in their “next token” prediction. Lower temp = more deterministic, Higher temp = a more creative output.
One approach taken was a “mimic” approach (task framing), where the models were prompted simply with “You are a clinical coder, consider the case note and assign the appropriate ICD codes”, which did allow the models tested to move in the right direction, but often made mistakes with mismatched codes and descriptions. The key part of this approach was the lack of examples given as a framework for the model to work with during the response.
The second approach was a “tree search” traversal information retrieval task. The models were given a case note and a description of the ICD code at each hierarchy level, and asked to determine if there were related mentions in the note.
- Clinical Coder: Manipulates the LLM to recall and apply its pre-trained knowledge of diseases and ICD codes, making a one-shot prediction. It’s like asking the LLM to “guess” the codes based on what it knows, which works better for common codes but struggles with rare ones or precise ontology details.
- Tree-Search: A full methodology where the LLM acts as a decision-maker to navigate the ICD hierarchy. It evaluates provided code descriptions at each level to decide which branches (nodes) to explore, systematically reaching leaf codes. This structured approach ensures coverage of rare codes and avoids reliance on the LLM’s imperfect memory.
Tree search appears to be far more computationally heavy, with multiple calls out to APIs for their decisions on which node to move to next. The algorithm denoted by the paper essentially traverses each node, and gives a fresh prompt and context to the LLM for each request. If we’re at the base node, we can return the ICD code, otherwise we continue to branch and traverse. It’s a much more systematic and definitely more robust approach than just kicking off a single prompt and expecting the result, especially in the rarer cases.
Some Q’s and future expansions:
- How can we reduce the amount of computation required? I think a new LLM call for each branch could be quite excessive. My solution would either be to distill or work with some kind of MOE model where instead of having multiple different experts for consultation, we have multiple of the same expert for almost chaining the next request to? so we can make a single request via API or inference gateway and then have our end result given?
- How does the model ensure determinism and confidence in it’s choice? What metrics allow the model to decide that it’s made the correct decision? Is this a weakpoint? What could be improved?
- Would this approach be coded up/integrated as part of a larger system for use by a doctor’s office etc? Can we apply this to other areas to free up a doctors time?
- How do you think these results would differ if run with newer models like Grok? Why?
- How can we improve either approach?
Grok did have a good chew on the MOTSE idea for an architecture, but it seems extremely tricky to implement! Some fine tuning wouldn’t be a bad idea either…
Clinical Coder Improvements:
- Prompt Engineering: Add examples or context about the ICD ontology to the prompt to reduce code-description mismatches
- Post-Processing: Validate outputs against the ICD ontology to correct mismatches
- Hybrid Approach: Combine with a lightweight classifier to pre-filter likely codes, guiding the LLM to focus on relevant concepts.
- Confidence Scores: Ask the LLM to provide confidence scores for each code, allowing filtering of low-confidence predictions.
Tree-Search Improvements:
- Batched Prompts: Evaluate multiple hierarchy levels or codes in a single prompt to reduce LLM calls, leveraging larger context windows.
- Confidence Integration: Request confidence scores in the prompt to prioritize high-confidence paths and flag uncertain ones for review.
- Backtracking Mechanism: Allow revisiting parent nodes if no leaf codes are found, reducing false negatives
- ICD Rules: Incorporate ICD coding rules to prevent mutually exclusive code predictions, using a rule-based filter post-LLM.
- Level-Specific Prompts: Tailor prompts for different hierarchy levels to improve accuracy.
- Ensemble or Multi-Model: Use multiple LLMs or prompt variations to aggregate decisions, improving robustness.
General Improvements:
- Pre-Filtering: Use a keyword-based or lightweight model to identify relevant chapters, reducing tree branches to explore.
- Caching: Store LLM responses for common case note patterns or code descriptions to reuse in similar cases.
- Human-in-the-Loop: Integrate a review interface for clinicians to validate or correct suggested codes, especially for low-confidence or rare cases.
- Fine-Tuning: If zero-shot is not strictly required, fine-tune the LLM on a small ICD dataset to improve accuracy without losing generalizability.